|
by Cliff Bueno de Mesquita Greetings and happy new year! To kick of the new year, I will continue with the methane cycling theme as I provide some information on the genus Methanolobus. Methanolobus is a genus of archaea in the family Methanosarcinaceae that can perform decomposition in anaerobic (low oxygen) environments. The lack of oxygen causes there to be different chemical reactions when these microbes eat carbon, compared to when, for example, humans eat carbon in the form of food. In anaerobic decomposition, methane is produced from either acetate, carbon dioxide and hydrogen, or various methyl (-- CH3) containing compounds. Generally, the family Methanosarcinaceae and genus Methanolobus are interesting because they are capable of using a variety of compounds and pathways to create methane (Oren, 2014). However, some species of Methanolobus have been reported to grow strictly on methyl compounds such as methanol and trimethylamine (König & Stetter, 1982). They have been found in lakes (Chen et al., 2018) and wetlands (Zhang et al., 2008) as well as a variety of niche environments such as coal seams (Doerfert et al., 2009) and oil wells (Ni & Boone, 1991). A Methanolobus genome was recently isolated from a former industrial salt pond in the San Francisco Bay. Industrial salt production involves creating a pond that is disconnected hydrologically so that water can evaporate and increase the salt concentration. The ponds are shallow and experience high UV radiation and high temperatures. Sadly, the majority of wetlands in the Bay Area were converted to industrial salt production. Even after the salt production stops, these areas can still have really elevated salt concentrations (5 times saltier than the sea!) as well as high sulfate levels. A new study that I’m involved in is reporting elevated methane emissions from these former industrial salt ponds, in part due to the activity of organisms such as Methanolobus. The good news is that efforts to restore these ponds back to their previous natural wetland state can change the microbial composition and lower the methane emissions. 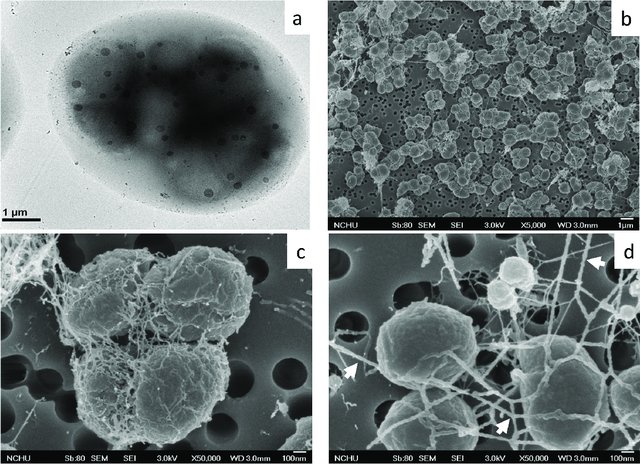 Figure 1. 1. Morphology of Methanolobus strain YSF-03T . (a) The negatively stained cells under transmission electron microscope JEM1400 (JEOL). (b)–(d) Cells under scanning electron microscope JSM-7401F (JEOL). Bars, (a) and (b), 1 µm; (c) and (d), 100 nm. The white arrows indicate the cannula-like structures. From Chen et al. 2018. Methanolobus are halophilic organisms, meaning “salt-loving”. Hence their presence in the former industrial salt ponds where salinity is still high. They’ve also been reported as abundant on hypersaline (super salty!) microbe mats in Baja California (Orphan et al., 2008). Some species can also be considered psychrophilic “cold-loving” (Zhang et al., 2008) but the genus as a whole can grow at a range of temperatures. In our study, the abundance of this organism was positively correlated with methane. We also found mcrA, mcrB, and mcrG genes in its genome which are the genes known to catalyze the final reaction in methanogenesis!
References: Chen, S.-C., Huang, H.-H., Lai, M.-C., Weng, C.-Y., Chiu, H.-H., Tang, S.-L., Rogozin, D. Y., & Degermendzhy, A. G. (2018). Methanolobus psychrotolerans sp. Nov., a psychrotolerant methanoarchaeon isolated from a saline meromictic lake in Siberia. International Journal of Systematic and Evolutionary Microbiology, 68(4), 1378–1383. https://doi.org/10.1099/ijsem.0.002685 Doerfert, S. N., Reichlen, M., Iyer, P., Wang, M., & Ferry, J. G. (2009). Methanolobus zinderi sp. Nov., a methylotrophic methanogen isolated from a deep subsurface coal seam. International Journal of Systematic and Evolutionary Microbiology, 59(5), 1064–1069. https://doi.org/10.1099/ijs.0.003772-0 König, H., & Stetter, K. O. (1982). Isolation and characterization of Methanolobus tindarius, sp. Nov., a coccoid methanogen growing only on methanol and methylamines. Zentralblatt Für Bakteriologie Mikrobiologie Und Hygiene: I. Abt. Originale C: Allgemeine, Angewandte Und Ökologische Mikrobiologie, 3(4), 478–490. https://doi.org/10.1016/S0721-9571(82)80005-7 Ni, S., & Boone, D. R. (1991). Isolation and Characterization of a Dimethyl Sulfide-Degrading Methanogen, Methanolobus siciliae HI350, from an Oil Well, Characterization of M. siciliae T4/MT, and Emendation of M. siciliae. International Journal of Systematic and Evolutionary Microbiology, 41(3), 410–416. https://doi.org/10.1099/00207713-41-3-410 Oren, A. (2014). The Family Methanosarcinaceae. In E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, & F. Thompson (Eds.), The Prokaryotes: Other Major Lineages of Bacteria and The Archaea (pp. 259–281). Springer. https://doi.org/10.1007/978-3-642-38954-2_408 Orphan, V. J., Jahnke, L. L., Embaye, T., Turk, K. A., Pernthaler, A., Summons, R. E., & Marais, D. J. D. (2008). Characterization and spatial distribution of methanogens and methanogenic biosignatures in hypersaline microbial mats of Baja California. Geobiology, 6(4), 376–393. https://doi.org/10.1111/j.1472-4669.2008.00166.x Zhang, G., Jiang, N., Liu, X., & Dong, X. (2008). Methanogenesis from Methanol at Low Temperatures by a Novel Psychrophilic Methanogen, “Methanolobus psychrophilus” sp. Nov., Prevalent in Zoige Wetland of the Tibetan Plateau. Applied and Environmental Microbiology, 74(19), 6114–6120. https://doi.org/10.1128/AEM.01146-08
0 Comments
Leave a Reply. |
AuthorVarious lab members contribute to the MoM Blog Archives
October 2023
Categories |

 RSS Feed
RSS Feed
